The XTAG tools are installed in the AFS. To get them to work on your account, you need to set the relevant paths:
$ xemacs ~/.bashrc
$ test -r /afs/sfs/lehre/wmaier/tag-seminar/.bashrc && . /afs/sfs/lehre/wmaier/tag-seminar/.bashrcThe referenced file contains all paths you need.
$ source ~/.bashrc
$ which runparserIts result should be
/afs/sfs/lehre/wmaier/xtag/lem-0.14.0/bin/runparser
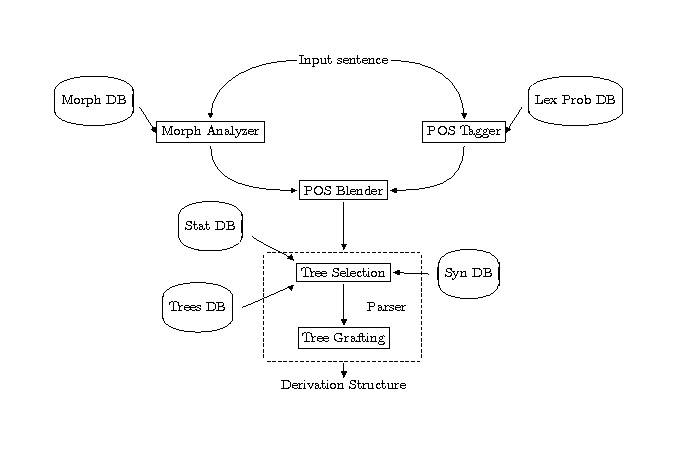
Morphological Analyzer and Morph Database: Consists of appr. 317000 inflected items derived from over 90000 stems. Returns root form, POS, and inflectional information.
We will take a look at the maintenance interface of the morphological database. It allows to add, edit or view entries of the morphological database. Please type
$ cp /afs/sfs/lehre/wmaier/xtag/tools/morph-1.5/data/morph_english.db ~/ $ cd ~/ $ xmdbm -db morph_english.dband lookup some interesting words:
You might want to delete the morphology database after using it:
$ rm ~/morph_english.db
The basis for tree selection of the parser is the presence of a POS tag on every word of the input. For those lexical items that cannot be assigned a POS tag via the Morphological Analyzer, the POS tagger is used. The POS Blender makes the final decision about the POS tag a word receives. It uses the output of the POS tagger as a filter on the output of the morphological analyzer. Any words that are not found in the morphological database are assigned the POS given by the tagger.
Tree Database: 1004 trees, divided into 53 tree families and 221 individual trees. The tree families represent subcategorization frames; the trees in a tree-family would be related to each other transformationally in a movement-based approach.
Syntactic Database: More than 30000 entries. Each entry consists of: uninflected form of the word, POS, list of trees or tree-families associated with the word, and a list of feature equations that capture lexical idiosyncrasies.
We will take a look at the maintenance interface of the syntatic database. Please run
$ syneditand load the following file:
/afs/sfs/lehre/wmaier/tag-seminar/xtag/syntax.flatThe interface can now be used to search the database or to view or edit entries.
Various tools for the access to the two databases are available.
$ xtag.show english [regexp]
xtag.show can be used to view individual trees from the grammar.
Any regular expression xtag.show english ^betaN[0-9]*
will display all the relative clause
trees.
$ xtag.show.fam english [regexp1] [regexp2]xtag.show.fam can be used to view trees in families from the grammar. Any tree family that matches [regexp1] and then each tree in that family which matches [regexp2] is displayed. e.g.
$ xtag.show.fam english ^Tnx0Vnx1$ ^alphaW[0-1]*will display all wh-extraction trees from the transitive tree family
$ xtag.show.word english [word] [regexp]xtag.show.word can be used to view all trees lexicalized by [word]. In addition, the list of all such trees can be filtered by using the optional [regexp] parameter. Use ".*" if you want to see all selected trees. e.g.
$ xtag.show.word english aim "for"will show only the trees that are anchored by the word "aim" and coanchored by the preposition "for". To see all trees for "aim", run the command
$ xtag.show.word english aim ".*". To see a transitive and an intransitive version of an elementary tree for bought, run
$ xtag.show.word english bought "alphanx0Vnx1\[bought\]" $ xtag.show.word english bought "alphanx0Vnx2nx1\[bought\]"
$ runparser /afs/sfs/lehre/wmaier/tag-seminar/testfile > outfile $ less outfile
Parses (derivation trees and derived trees) can be extracted from the forest with print_deriv. Try
$ cat outfile | print_deriv -p > outfile.derived $ cat outfile | print_deriv -d > outfile.derivation
The first command extracts all derived trees, the second command extracts all derivation trees. Take a look at the new files. They contain trees written in a bracketed notation. You can view them graphically with showtrees. (Navigation: right mouse button: next tree, left mouse button: previous tree)
$ cat outfile.xxx | showtrees
Of course you can run all at once using pipes:
$ runparser /afs/sfs/lehre/wmaier/tag-seminar/testfile | print_deriv -p | showtrees
$ runparser +c /afs/sfs/lehre/wmaier/tag-seminar/testfile | print_deriv -f | showtrees
A window should pop up with parses that have had successful unifications. Pressing 'f' in the window shows you the feature structures of each tree.
To unify feature structures after parsing:
$ runparser +u outfile | print_deriv -f | showtrees
$ xtag.browserand load one of the parse forest file you have produced before.